Minor Flood Frequency and Duration#
In this notebook we will plot two indicators concerning flooding at the Malakal tide gauge, after first taking a general look at the type of data we are able to plot from the UH Sea Level Center. These indicators are based on a ‘flooding’ threshold, using relative sea level.
Setup#
We first need to import the necessary libraries, access the data, and make a quick plot to ensure we will be analyzing the right thing.
Import necessary libraries.#
# Standard libraries
import os
import os.path as op
import datetime
from pathlib import Path
import sys
# Data manipulation libraries
import numpy as np
import pandas as pd
import xarray as xr
# Data analysis libraries
import scipy.stats as stats
# Visualization libraries
import matplotlib.pyplot as plt
import seaborn as sns
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from matplotlib.colors import Normalize
from urllib.request import urlretrieve #used for downloading files
# Miscellaneous
from myst_nb import glue # used for figure numbering when exporting to LaTeX
sys.path.append("../../../functions")
from data_downloaders import download_oni_index
Next, we’ll establish our directory for saving and reading data, and our output files.
Caution
You will need to change the variable for “output_dir” to your own output path to save figures and tables to a place of your choosing.
data_dir = Path('../../../data')
path_figs = "../../../matrix_cc/figures"
data_dir = Path(data_dir,'sea_level')
#CHANGE THIS TO YOUR PATH!!
output_dir = Path('/Users/laurac/Library/Mobile Documents/com~apple~CloudDocs/Projects/CC_indicators/CC_indicators/data/') # EDIT THIS TO YOUR PATH
# Create the output directory if it does not exist
output_dir.mkdir(exist_ok=True)
data_dir.mkdir(exist_ok=True)
Retrieve the Tide Station(s) Data Set(s)#
Next, we’ll access data from the UHSLC. The Malakala tide gauge has the UHSLC ID: 7. We will import the netcdf file into our current data directory, and take a peek at the dataset. We will also import the datums for this location.
uhslc_id = 7
# download the hourly data
fname = f'h{uhslc_id:03}.nc' # h for hourly, d for daily
url = "https://uhslc.soest.hawaii.edu/data/netcdf/fast/hourly/"
path = os.path.join(data_dir, fname)
if not os.path.exists(path):
urlretrieve(os.path.join(url, fname), path)
rsl = xr.open_dataset(data_dir / fname)
# remove the trailing zero from record_id. This zero is added to the record_id to make it unique if the station has multiple entries
rsl['record_id'] =(rsl['record_id']/10).astype(int)
rsl
<xarray.Dataset> Size: 6MB
Dimensions: (record_id: 1, time: 486849)
Coordinates:
* time (time) datetime64[ns] 4MB 1969-05-18T15:00:00 ... 2...
* record_id (record_id) int64 8B 7
Data variables:
sea_level (record_id, time) float32 2MB ...
lat (record_id) float32 4B ...
lon (record_id) float32 4B ...
station_name (record_id) |S7 7B ...
station_country (record_id) |S5 5B ...
station_country_code (record_id) float32 4B ...
uhslc_id (record_id) int16 2B ...
gloss_id (record_id) float32 4B ...
ssc_id (record_id) |S4 4B ...
last_rq_date (record_id) datetime64[ns] 8B ...
Attributes:
title: UHSLC Fast Delivery Tide Gauge Data (hourly)
ncei_template_version: NCEI_NetCDF_TimeSeries_Orthogonal_Template_v2.0
featureType: timeSeries
Conventions: CF-1.6, ACDD-1.3
date_created: 2025-01-27T14:35:24Z
publisher_name: University of Hawaii Sea Level Center (UHSLC)
publisher_email: philiprt@hawaii.edu, markm@soest.hawaii.edu
publisher_url: http://uhslc.soest.hawaii.edu
summary: The UHSLC assembles and distributes the Fast Deli...
processing_level: Fast Delivery (FD) data undergo a level 1 quality...
acknowledgment: The UHSLC Fast Delivery database is supported by ...and we’ll save a few variables that will come up later for report generation.
Show code cell source
station = rsl['station_name'].values[0].decode('utf-8')
country = rsl['station_country'].values[0].decode('utf-8')
startDateTime = str(rsl.time.values[0])[:10]
endDateTime = str(rsl.time.values[-1])[:10]
station_name = station + ', ' + country
glue("station",station,display=False)
glue("country",country, display=False)
glue("startDateTime",startDateTime, display=False)
glue("endDateTime",endDateTime, display=False)
Set the Datum to MHHW#
In this example, we will set the datum to MHHW. This can be hard coded, or we can write a script to read in the station datum information from UHSLC, below.
def get_MHHW_uhslc_datums(id, datumname):
url = 'https://uhslc.soest.hawaii.edu/stations/TIDES_DATUMS/fd/LST/fd'+f'{int(id):03}'+'/datumTable_'+f'{int(id):03}'+'_mm_GMT.csv'
datumtable = pd.read_csv(url)
datum = datumtable[datumtable['Name'] == datumname]['Value'].values[0]
# ensure datum is a float
datum = float(datum)
return datum, datumtable
# extract the given datum from the dataframe
datumname = 'MHHW'
datum, datumtable = get_MHHW_uhslc_datums(uhslc_id, datumname)
rsl['datum'] = datum # already in mm
rsl['sea_level_mhhw'] = rsl['sea_level'] - rsl['datum']
# assign units to datum and sea level
rsl['datum'].attrs['units'] = 'mm'
rsl['sea_level_mhhw'].attrs['units'] = 'mm'
glue("datum", datum, display=False)
glue("datumname", datumname, display=False)
datumtable
Show code cell output
| Name | Value | Description | |
|---|---|---|---|
| 0 | Status | 23-Aug-2025 | Processing Date |
| 1 | Epoch | 01-Jan-1983 to 31-Dec-2001 | Tidal Datum Analysis Period |
| 2 | MHHW | 2160 | Mean Higher-High Water (mm) |
| 3 | MHW | 2088 | Mean High Water (mm) |
| 4 | MTL | 1530 | Mean Tide Level (mm) |
| 5 | MSL | 1532 | Mean Sea Level (mm) |
| 6 | DTL | 1457 | Mean Diurnal Tide Level (mm) |
| 7 | MLW | 973 | Mean Low Water (mm) |
| 8 | MLLW | 754 | Mean Lower-Low Water (mm) |
| 9 | STND | 0 | Station Datum (mm) |
| 10 | GT | 1406 | Great Diurnal Range (mm) |
| 11 | MN | 1114 | Mean Range of Tide (mm) |
| 12 | DHQ | 72 | Mean Diurnal High Water Inequality (mm) |
| 13 | DLQ | 219 | Mean Diurnal Low Water Inequality (mm) |
| 14 | HWI | Unavailable | Greenwich High Water Interval (in hours) |
| 15 | LWI | Unavailable | Greenwich Low Water Interval (in hours) |
| 16 | Maximum | 2785 | Highest Observed Water Level (mm) |
| 17 | Max Time | 23-Jun-2013 22 | Highest Observed Water Level Date and Hour (GMT) |
| 18 | Minimum | -34 | Lowest Observed Water Level (mm) |
| 19 | Min Time | 19-Jan-1992 16 | Lowest Observed Water Level Date and Hour (GMT) |
| 20 | HAT | 2567 | Highest Astronomical Tide (mm) |
| 21 | HAT Time | 27-Aug-1988 22 | HAT Date and Hour (GMT) |
| 22 | LAT | 175 | Lowest Astronomical Tide (mm) |
| 23 | LAT Time | 31-Dec-1986 16 | LAT Date and Hour (GMT) |
| 24 | LEV | 1644 | Switch 1 (mm) |
| 25 | LEVB | 1541 | Switch 2 (mm) |
Assess Station Data Quality for the POR (1983-2022)#
To do this, we’ll plot all the sea level data to make sure our data looks correct, and then we’ll truncate the data set to the time period of record (POR).
fig, ax = plt.subplots(sharex=True)
fig.autofmt_xdate()
ax.plot(rsl.time.values,rsl.sea_level_mhhw.T.values/10)
ax.set_ylabel(f'Sea Level (cm, {datumname})') #divide by 10 to convert to cm
glue("TS_full_fig",fig,display=False)
Show code cell output
Identify epoch for the flood frequency analysis#
Now, we’ll calculate trend starting from the beginning of the tidal datum analysis period epoch to the last time processed. The epoch information is given in the datums table.
#get epoch start time from the epoch in the datumtable
epoch_times = datumtable[datumtable['Name'] == 'Epoch']['Value'].values[0]
#parse epoch times into start time
epoch_start = epoch_times.split(' ')[0]
epoch_start = datetime.datetime.strptime(epoch_start, '%d-%b-%Y')
# and for now, end time the processind end time
epoch_end = epoch_times.split(' ')[2]
epoch_end = datetime.datetime.strptime(epoch_end, '%d-%b-%Y')
# last date is rsl['last_rq_date'].values
data_end = rsl['time'].values[-1]
data_end = pd.to_datetime(data_end)
# start the data at year before epoch_start year on May 1st
data_start = pd.Timestamp(year=epoch_start.year-1, month=5, day=1)
data_start = pd.to_datetime(data_start)
# end the data at April 30th of the year of the last data request
data_end = pd.Timestamp(year=data_end.year, month=4, day=30)
data_end = pd.to_datetime(data_end)
hourly_data = rsl.sel(dict(time=slice(data_start, data_end)))
hourly_data
glue("startDataDateTime",data_start.strftime('%Y-%m-%d'), display=False)
glue("endDataDateTime",data_end.strftime('%Y-%m-%d'), display=False)
glue("startEpochDateTime",epoch_start.strftime('%Y-%m-%d'), display=False)
glue("endEpochDateTime",epoch_end.strftime('%Y-%m-%d'), display=False)
and plot the hourly time series
fig, ax = plt.subplots(sharex=True)
fig.autofmt_xdate()
ax.plot(hourly_data.time.values,hourly_data.sea_level_mhhw.T.values/10) #divide by 10 to convert to cm
ax.set_ylabel(f'Sea Level (cm, {datumname})')
glue("TS_epoch_fig",fig,display=False)
Show code cell output
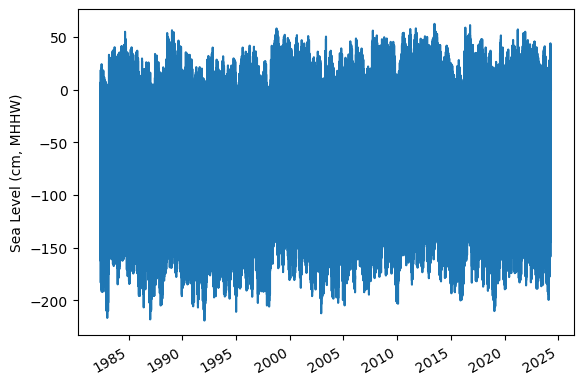

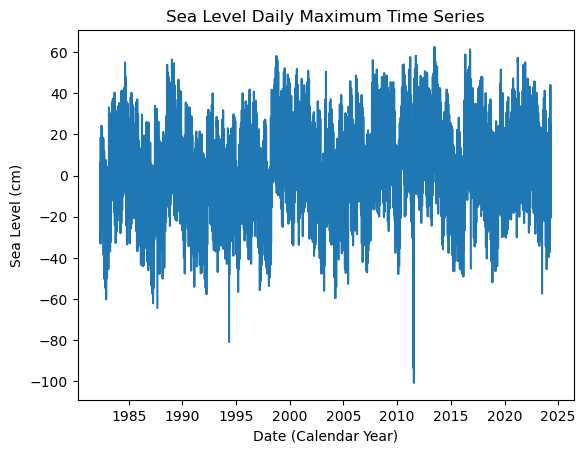
Fig. 5 Full time series at Malakal,Palau tide gauge for the entire record from 1982-05-01 to 2024-04-30. Note that the sea level is plotted in units of cm, relative to MHHW.#
Assign a Threshold#
The threshold used here to determine a flood event is 30 cm above MHHW.
Note
Change the threshold and see how things change!
threshold = 30 # in cm
glue("threshold",threshold,display=False)
Calculate and Plot Flood Frequency#
To analyze flood frequency, we will look for daily maximum sea levels for each day in our dataset, following and others. Then, we can group our data by year and month to visualize temporal patterns in daily SWL exceedance.
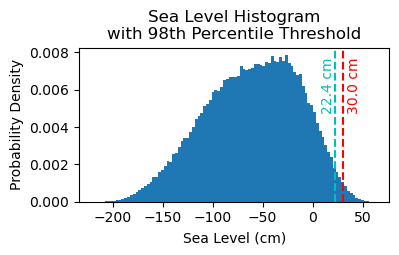
Fig. 6 Histogram of daily maximum water levels at Malakal,Palau tide gauge for the entire record from 1982-05-01 to 2024-04-30, relative to MHHW. The dashed red line indicates the chosen threshold of 30 cm.#
# Resample the hourly data to daily maximum sea level
SL_daily_max = hourly_data.resample(time='D').max()
static_vars = hourly_data[['station_name', 'station_country','station_country_code', 'datum','lat', 'lon','uhslc_id','gloss_id','ssc_id','last_rq_date']]
SL_daily_max = SL_daily_max.assign(static_vars)
SL_daily_max
<xarray.Dataset> Size: 307kB
Dimensions: (time: 15341, record_id: 1)
Coordinates:
* record_id (record_id) int64 8B 7
* time (time) datetime64[ns] 123kB 1982-05-01 ... 2024-04-30
Data variables:
sea_level (time, record_id) float32 61kB 1.881e+03 ... 2.161e+03
lat (record_id) float32 4B 7.33
lon (record_id) float32 4B 134.5
station_name (record_id) |S7 7B b'Malakal'
station_country (record_id) |S5 5B b'Palau'
station_country_code (record_id) float32 4B 585.0
uhslc_id (record_id) int16 2B 7
gloss_id (record_id) float32 4B 120.0
ssc_id (record_id) |S4 4B b'mala'
last_rq_date (record_id) datetime64[ns] 8B 2018-12-31T22:59:59.9...
datum float64 8B 2.16e+03
sea_level_mhhw (time, record_id) float64 123kB -279.0 -161.0 ... 1.0
Attributes:
title: UHSLC Fast Delivery Tide Gauge Data (hourly)
ncei_template_version: NCEI_NetCDF_TimeSeries_Orthogonal_Template_v2.0
featureType: timeSeries
Conventions: CF-1.6, ACDD-1.3
date_created: 2025-01-27T14:35:24Z
publisher_name: University of Hawaii Sea Level Center (UHSLC)
publisher_email: philiprt@hawaii.edu, markm@soest.hawaii.edu
publisher_url: http://uhslc.soest.hawaii.edu
summary: The UHSLC assembles and distributes the Fast Deli...
processing_level: Fast Delivery (FD) data undergo a level 1 quality...
acknowledgment: The UHSLC Fast Delivery database is supported by ...# make a new figure that is 15 x 5
fig, ax = plt.subplots(sharex=True)
plt.plot(SL_daily_max.time.values, SL_daily_max.sea_level_mhhw.values/10)
plt.xlabel('Date (Calendar Year)')
plt.ylabel('Sea Level (cm)')
plt.title('Sea Level Daily Maximum Time Series')
Text(0.5, 1.0, 'Sea Level Daily Maximum Time Series')
# Make a pdf of the data with 95th percentile threshold
sea_level_data_cm = hourly_data['sea_level_mhhw'].values/10 # convert to cm
#remove nans
sea_level_data_cm = sea_level_data_cm[~np.isnan(sea_level_data_cm)]
fig, ax = plt.subplots(figsize=(4,2))
ax.hist(sea_level_data_cm, bins=100, density=True, label='Sea Level Data')
#get 95th percentile
threshold98 = np.percentile(sea_level_data_cm, 98)
ax.axvline(threshold, color='r', linestyle='--', label='Threshold: {:.4f} cm'.format(threshold))
ax.axvline(threshold98, color='c', linestyle='--', label='Threshold: {:.4f} cm'.format(threshold98))
ax.set_xlabel('Sea Level (cm)')
ax.set_ylabel('Probability Density')
# make the title two lines
ax.set_title('Sea Level Histogram\nwith 98th Percentile Threshold')
# add label to dashed line
# get value of middle of y-axis for label placement
ymin, ymax = ax.get_ylim()
yrange = ymax - ymin
y_middle = ymin + 0.75*yrange
ax.text(threshold+4, y_middle, '{:.1f} cm'.format(threshold), rotation=90, va='center', ha='left', color='r')
ax.text(threshold98, y_middle, '{:.1f} cm'.format(threshold98), rotation=90, va='center', ha='right', color='c')
glue("histogram_fig", fig, display=False)
Show code cell output

# Find all days where sea level exceeds the threshold
flood_days_df = SL_daily_max.to_dataframe().reset_index()
flood_days_df['year_storm'] = flood_days_df['time'].dt.year
flood_days_df.loc[flood_days_df['time'].dt.month > 4, 'year_storm'] = flood_days_df['time'].dt.year + 1
#filter flood hours
flood_days_df = flood_days_df[flood_days_df['sea_level_mhhw'] > threshold*10]
flood_days_per_year = flood_days_df.groupby('year_storm').size().reset_index(name='flood_days_count')
all_years = pd.DataFrame({'year_storm': range(flood_days_df['year_storm'].min(), flood_days_df['year_storm'].max() + 1)})
flood_days_per_year = all_years.merge(flood_days_per_year, on='year_storm', how='left').fillna(0)
flood_days_per_year['flood_days_count'] = flood_days_per_year['flood_days_count'].astype(int)
flood_hours_df = hourly_data.to_dataframe().reset_index()
flood_hours_df['year_storm'] = flood_hours_df['time'].dt.year
flood_hours_df.loc[flood_hours_df['time'].dt.month > 4, 'year_storm'] = flood_hours_df['time'].dt.year + 1
#filter flood hours
flood_hours_df = flood_hours_df[flood_hours_df['sea_level_mhhw'] > threshold*10]
flood_hours_per_year = flood_hours_df.groupby('year_storm').size().reset_index(name='flood_hours_count')
all_years = pd.DataFrame({'year_storm': range(flood_hours_df['year_storm'].min(), flood_hours_df['year_storm'].max() + 1)})
flood_hours_per_year = all_years.merge(flood_hours_per_year, on='year_storm', how='left').fillna(0)
flood_hours_per_year['flood_hours_count'] = flood_hours_per_year['flood_hours_count'].astype(int)
Let’s have a quick look at the data in flood_hours_per_year:
# show the first few rows of the flood_days_per_year dataframe
flood_days_per_year.head()
| year_storm | flood_days_count | |
|---|---|---|
| 0 | 1983 | 1 |
| 1 | 1984 | 33 |
| 2 | 1985 | 40 |
| 3 | 1986 | 9 |
| 4 | 1987 | 0 |
# add nino
p_data = 'https://psl.noaa.gov/data/correlation/oni.data'
oni = download_oni_index(p_data)
/Users/laurac/Library/Mobile Documents/com~apple~CloudDocs/Projects/CC_indicators/CC_indicators/notebooks_historical/ocean/1_sea_level/../../../functions/data_downloaders.py:242: FutureWarning: The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
oni = pd.read_csv(
# add column for oni values to flood_days_per_year, with average ONI value for the storm year
# first establish storm year for each , given that the date is the index
oni['year_storm'] = oni.index.year
# if the month is less than 5, then the storm year is the year before
oni.loc[oni.index.month > 4, 'year_storm'] = oni.loc[oni.index.month > 4, 'year_storm'] + 1
# group by storm year and get the mean ONI value
oni_year = oni.groupby('year_storm').mean()
# merge the oni values with the flood_days_per_year dataframe
flood_days_per_year = flood_days_per_year.merge(oni_year, on='year_storm', how='left')
def detect_enso_events(oni_df):
"""
Detects El Niño and La Niña events within a storm year based on the ONI index.
An El Niño event is defined as having 5 consecutive months with ONI > 0.5.
A La Niña event is defined as having 5 consecutive months with ONI < -0.5.
Parameters:
oni_df (pd.DataFrame): DataFrame containing the ONI index with a datetime index.
Returns:
pd.DataFrame: DataFrame with columns 'year_storm' and 'event' indicating the type of event ('El Niño', 'La Niña', or 'Neutral').
"""
oni_df['event'] = 'Neutral'
oni_df['year_storm'] = oni_df.index.year
oni_df.loc[oni_df.index.month > 4, 'year_storm'] += 1
for year in oni_df['year_storm'].unique():
yearly_data = oni_df[oni_df['year_storm'] == year]['ONI']
for i in range(len(yearly_data) - 4):
if all(yearly_data.iloc[i:i+5] > 0.5):
oni_df.loc[oni_df['year_storm'] == year, 'event'] = 'El Niño'
break
elif all(yearly_data.iloc[i:i+5] < -0.5):
oni_df.loc[oni_df['year_storm'] == year, 'event'] = 'La Niña'
break
return oni_df[['year_storm', 'event']].drop_duplicates().reset_index(drop=True)
# Example usage:
# oni_df = download_oni_index(p_data) # Assuming this function is already defined and returns a DataFrame with ONI index
enso_events = detect_enso_events(oni)
# show the first few rows of the enso_events dataframe
enso_events.head()
| year_storm | event | |
|---|---|---|
| 0 | 1951 | Neutral |
| 1 | 1952 | El Niño |
| 2 | 1953 | Neutral |
| 3 | 1954 | El Niño |
| 4 | 1955 | La Niña |
# add enso events to flood_days_per_year
flood_days_per_year = flood_days_per_year.merge(enso_events, on='year_storm', how='left')
flood_days_per_year.head()
| year_storm | flood_days_count | ONI | event | |
|---|---|---|---|---|
| 0 | 1983 | 1 | 1.510833 | El Niño |
| 1 | 1984 | 33 | -0.246667 | Neutral |
| 2 | 1985 | 40 | -0.643333 | La Niña |
| 3 | 1986 | 9 | -0.434167 | Neutral |
| 4 | 1987 | 0 | 0.745000 | El Niño |
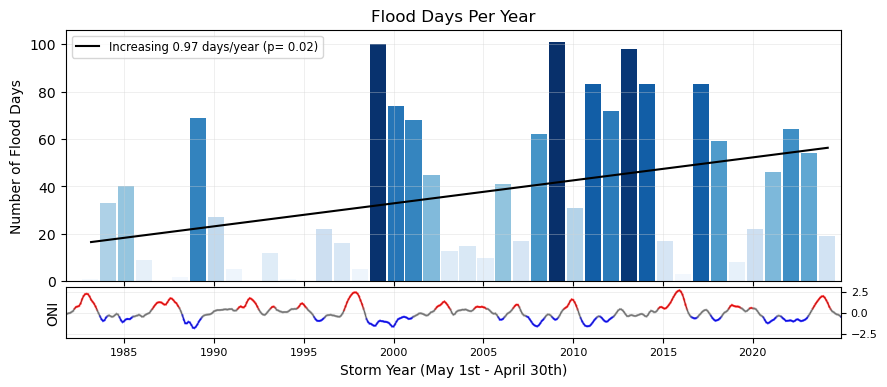
Plot Flood Frequency Counts#
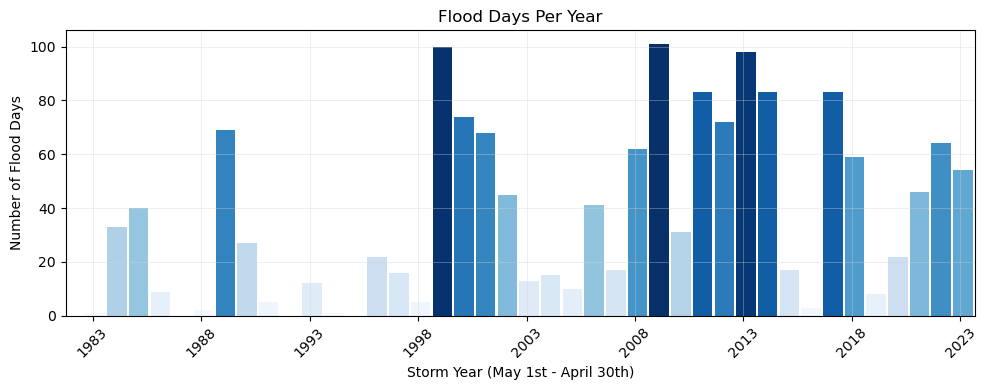
The flood frequency counts are defined as the number of time periods that exceed a given threshold within a year. This plot follows .
# Create two subplots sharing the same x-axis
fig, (ax1, ax2) = plt.subplots(
2, 1, figsize=(10, 4), sharex=True, gridspec_kw={"height_ratios": [5, 1],"hspace": 0.04, }
)
# Plot the bar chart for flood days on the first subplot
x_values = flood_days_per_year['year_storm'] # Assuming this is already aligned to storm years
x_values_offset = x_values - 4/12 # Shift x-values to align with the storm years
y_values = flood_days_per_year['flood_days_count']
bar_width = 0.9
flooding_colors = sns.color_palette("Blues", as_cmap=True)
norm = Normalize(vmin=min(y_values), vmax=max(y_values))
colors = flooding_colors(norm(y_values))
ax1.bar(x_values_offset, y_values, width=bar_width, color=colors, align='edge')
ax1.set_ylabel('Number of Flood Days')
ax1.set_title('Flood Days Per Year')
ax1.grid(color='lightgray', linestyle='-', linewidth=0.5, alpha=0.5)
# Plot the ONI index on the second subplot
oni['year_float'] = oni.index.year + (oni.index.month - 1) / 12
# shifted x-axis to align with flood days histogram storm years
# x_offset = flood_days_per_year['year_storm'].min() + 4/12
ax2.plot(oni['year_float'], oni['ONI'], color='k',linewidth=0.5, label='ONI')
ax2.set_ylabel('ONI')
ax2.set_xlabel('Storm Year (May 1st - April 30th)')
ax2.grid(color='lightgray', linestyle='-', linewidth=0.5, alpha=0.5)
ax2.set_ylim(-3, 3)
ax1.set_xlim(x_values_offset.min()-bar_width, x_values.max()+bar_width)
ax2.set_xlim(x_values_offset.min()-bar_width, x_values.max()+bar_width)
# Define thresholds for ONI values
el_nino_threshold = 0.5
la_nina_threshold = -0.5
# Loop through the data to plot segments with different colors
for i in range(len(oni) - 1):
x = oni['year_float'].iloc[i:i+2]
y = oni['ONI'].iloc[i:i+2]
if y.mean() >= el_nino_threshold:
color = 'red'
elif y.mean() <= la_nina_threshold:
color = 'blue'
else:
color = 'gray'
ax2.plot(x, y, color=color, linewidth=1.5, alpha=0.5)
# put y-axis labels on the right side for ax2
ax2.yaxis.tick_right()
# set font size for tick labels
ax2.tick_params(axis='both', which='major', labelsize=8)
# Add the trendline to the first subplot
# Aggregating data and fitting the trendline
slope, intercept, r_value, p_value, std_err = stats.linregress(x_values_offset+0.5, y_values)
trendDays = intercept + slope * (x_values_offset+0.5)
trendLabel = 'Increasing {:.2f} days/year (p= {:.2f})'.format(slope,p_value)
if p_value < 0.05:
linestyleTrend = '-'
else:
linestyleTrend = '--'
ax1.plot(x_values_offset+0.5, trendDays, color='black', linestyle=linestyleTrend, label=trendLabel)
# Adding the legend
ax1.legend(loc='upper left', fontsize='small')
# # save the trendline values
glue("day_per_year_trend",slope,display=False)
glue("p_value_days",p_value,display=False)
# # save the figure
glue("fig-threshold_counts_enso", fig, display=False)
# save to output directory
fig.savefig(output_dir / "SL_FloodFrequency_threshold_counts_days.png", dpi=300, bbox_inches='tight')

Here is a more simplified plot of the same thing.
x_values = flood_days_per_year['year_storm'] # Assuming this is already aligned to storm years
y_values = flood_days_per_year['flood_days_count']
bar_width = 0.9 # Adjust the width of the bars
# Offset x-values by 4/12 (4 months = May start)
x_values_offset = x_values - 4/12
# Plot the bar chart
fig, ax = plt.subplots(figsize=(10, 4))
ax.bar(x_values_offset, y_values, width=bar_width, color=colors, align='edge')
# Add gridlines for better readability
ax.grid(color='lightgray', linestyle='-', linewidth=0.5, alpha=0.5)
# Add labels and title
ax.set_xlabel('Storm Year (May 1st - April 30th)')
ax.set_ylabel('Number of Flood Days')
ax.set_title('Flood Days Per Year')
# Adjust x-ticks to match storm years
ax.set_xticks(x_values[::5]) # Use storm years for ticks, spaced every 5 years
ax.set_xticklabels(x_values[::5].astype(int), rotation=45)
# Set x-limits to ensure all bars are visible
ax.set_xlim(x_values_offset.min() - bar_width, x_values_offset.max())
# Show the plot
plt.tight_layout()
plt.show()
Plot Flood Duration#
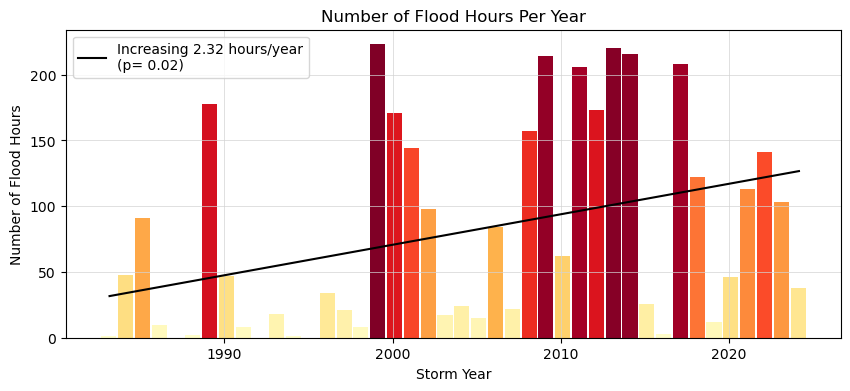
This next plot examines the average duration of flooding events as defined by the threshold. Note that in this calculation these hours need not be continuous (and thus any changes in the scale of the flooding in terms of time-extent is not readily apparent, only the yearly summation.)
# Adjusting the heatmap palette to improve readability
from matplotlib.colors import Normalize
adjusted_heatmap_palette = sns.color_palette("YlOrRd", as_cmap=True)
# Plot the bar chart
fig, ax = plt.subplots(figsize=(10, 4))
# Adding a light gray grid
ax.grid(color='lightgray', linestyle='-', linewidth=0.5)
y_values = flood_hours_per_year['flood_hours_count']
x_values = flood_hours_per_year['year_storm']
bar_width = 0.9
norm = Normalize(vmin=min(y_values), vmax=max(y_values))
colors = adjusted_heatmap_palette(norm(y_values))
# Offset x-values by 4/12 (4 months = May start)
x_values_offset = x_values - 4/12
ax.bar(x_values_offset, y_values, width=bar_width, color=colors, align='edge')
ax.set_xlabel('Storm Year')
ax.set_ylabel('Number of Flood Hours')
ax.set_title('Number of Flood Hours Per Year')
slope, intercept, r_value, p_value, std_err = stats.linregress(x_values_offset+0.5, y_values)
trendHours = intercept + slope * (x_values_offset+0.5)
trendLabel = 'Increasing {:.2f} hours/year\n(p= {:.2f})'.format(slope, p_value)
if p_value < 0.05:
linestyleTrend = '-'
else:
linestyleTrend = '--'
ax.plot(x_values_offset+0.5, trendHours, color='black', linestyle=linestyleTrend, label=trendLabel)
# Adding the legend
ax.legend(loc='upper left', fontsize='small')
# # save the trendline values
# glue("hours_per_year_trend",slope,display=False)
# glue("p_value_hours",p_value,display=False)
# Adding the legend
ax.legend(loc='upper left')
# glue("duration_fig", fig, display=False)
# save the figure
fig.savefig(output_dir / 'SL_FloodFrequency_threshold_counts_hours.png', bbox_inches='tight')
Show code cell output
Calculate the percent change over time#
Next we’ll calculate the percent change over the POR at the tide station/s, for both Frequency and Duration.
The next code cell fits a trend line to the flood days per year data and calculates the trend line. It then plots the data and the trend line in a figure, and calculates the percent change in flood days per year using the trend line. The same process is repeated for flood hours per year.
Now we’ll generate a table with this information, which will be saved as a .csv in the output directory specified at the top of this notebook.
# Calculate the total number of days/hours exceeding the threshold
total_flood_days = flood_days_per_year['flood_days_count'].sum()
total_flood_hours = flood_hours_per_year['flood_hours_count'].sum()
# Calculate the average number of flood days and hours per year
average_flood_days_per_year = total_flood_days / len(flood_days_per_year)
average_flood_hours_per_year = flood_hours_per_year['flood_hours_count'].mean()
# Find the maximum number of flood days in a single year
max_flood_days = flood_days_per_year['flood_days_count'].max()
max_flood_days_year = flood_days_per_year.loc[flood_days_per_year['flood_days_count'].idxmax(), 'year_storm']
# Find the maximum number of flood hours in a single year
max_flood_hours = flood_hours_per_year['flood_hours_count'].max()
max_flood_hours_year = flood_hours_per_year.loc[flood_hours_per_year['flood_hours_count'].idxmax(), 'year_storm']
# calculate the absolute percent change over the entire period using the first and last part of the trend
percent_change_days = (trendDays.iloc[-1]-trendDays.iloc[0])/trendDays.iloc[0]*100
percent_change_hours = (trendHours.iloc[-1]-trendHours.iloc[0])/trendHours.iloc[0]*100
# Create a dataframe for statistics
summary_stats = [
('Total Flood Days', int(total_flood_days)),
('Average Flood Days per Year', int(average_flood_days_per_year)),
('Max Flood Days in a Single Year', int(max_flood_days)),
('Year of Max Flood Days', int(max_flood_days_year)),
('Total Flood Hours', int(total_flood_hours)),
('Average Flood Hours per Year', int(average_flood_hours_per_year)),
('Max Flood Hours in a Single Year', int(max_flood_hours)),
('Year of Max Flood Hours', int(max_flood_hours_year)),
('Percent Change in Flood Days', int(percent_change_days)),
('Percent Change in Flood Hours', int(percent_change_hours))
]
# Convert the list of tuples to a pandas DataFrame
summary_stats_df = pd.DataFrame(summary_stats, columns=['Statistic', 'Value'])
# Display the DataFrame
summary_stats_df
| Statistic | Value | |
|---|---|---|
| 0 | Total Flood Days | 1530 |
| 1 | Average Flood Days per Year | 36 |
| 2 | Max Flood Days in a Single Year | 101 |
| 3 | Year of Max Flood Days | 2009 |
| 4 | Total Flood Hours | 3325 |
| 5 | Average Flood Hours per Year | 79 |
| 6 | Max Flood Hours in a Single Year | 223 |
| 7 | Year of Max Flood Hours | 1999 |
| 8 | Percent Change in Flood Days | 239 |
| 9 | Percent Change in Flood Hours | 300 |
Create a sub-annual table#
Here we’ll turn our ‘flooding days’ into a dataframe to look at monthly changes through time.
Show code cell source
import calendar
# Make a dataframe with years as rows and months as columns
df = SL_daily_max.to_dataframe().reset_index()
df['flood_day'] = df['sea_level_mhhw'] > threshold*10
# keep only the flood day and time columns
df = df[['time', 'flood_day']]
df['year'] = df['time'].dt.year
df['month'] = df['time'].dt.month
# adjust for storm year
df.loc[df['month'] > 4, 'year'] = df.loc[df['month'] > 4, 'year'] + 1
# drop original time column
df = df.drop(columns=['time'])
# sum the number of flood days in each month for each storm year
df = df.groupby(['year', 'month']).sum()
# pivot the table
df = df.pivot_table(index='year', columns='month', values='flood_day')
# remove first and last rows (partial years)
# df = df.iloc[1:-1]
# rename year to Storm Year
df.index.name = 'Storm Year'
#define reordered months (for storm year, May to April)
month_order = [5,6,7,8,9,10,11,12,1,2,3,4]
month_names = [calendar.month_abbr[i] for i in month_order]
# rename months to month names
df = df[month_order]
df.columns = month_names
# add a column for the total number of flood days in each year
df['Annual'] = df.sum(axis=1)
# add a row for the total number of flood days in each month, normalized the total flood days (last row, last column)
df.loc['Monthly Total (%)'] = df.sum()
df.loc['Monthly Total (%)'] = 100*df.loc['Monthly Total (%)']/df.loc['Monthly Total (%)', 'Annual']
station_name = SL_daily_max['station_name'].values[0].decode('utf-8') +', ' + SL_daily_max['station_country'].values[0].decode('utf-8')
tableTitle = station_name + ': Days Exceeding ' + str(threshold) + ' cm above MHHW'
# Apply background gradient and add a title with larger text
styled_df = df.style.background_gradient(
cmap='Purples', axis=None, subset=(df.index[:-1], df.columns[:-1])
).format("{:.0f}").background_gradient(
cmap='Grays', axis=None, subset=(df.index[-1], df.columns[:-1])
).format("{:.0f}").set_caption(tableTitle).set_table_styles([
{
'selector': 'caption',
'props': [('font-size', '16px'), ('margin-bottom', '10px')]
}
])
styled_df
| May | Jun | Jul | Aug | Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | Annual | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Storm Year | |||||||||||||
| 1983 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| 1984 | 1 | 5 | 5 | 4 | 3 | 2 | 1 | 0 | 2 | 3 | 1 | 6 | 33 |
| 1985 | 5 | 1 | 4 | 9 | 6 | 3 | 4 | 0 | 0 | 0 | 3 | 5 | 40 |
| 1986 | 3 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| 1987 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1988 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 1989 | 0 | 0 | 4 | 8 | 6 | 7 | 4 | 3 | 17 | 7 | 7 | 6 | 69 |
| 1990 | 4 | 0 | 2 | 6 | 5 | 4 | 4 | 0 | 0 | 0 | 0 | 2 | 27 |
| 1991 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| 1992 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1993 | 1 | 2 | 0 | 1 | 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
| 1994 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1995 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1996 | 0 | 0 | 1 | 3 | 0 | 2 | 4 | 0 | 0 | 1 | 0 | 11 | 22 |
| 1997 | 0 | 0 | 2 | 5 | 3 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 16 |
| 1998 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 |
| 1999 | 6 | 5 | 12 | 13 | 12 | 13 | 7 | 6 | 11 | 5 | 7 | 3 | 100 |
| 2000 | 5 | 7 | 14 | 11 | 10 | 10 | 6 | 4 | 3 | 4 | 0 | 0 | 74 |
| 2001 | 1 | 1 | 8 | 15 | 12 | 7 | 3 | 0 | 1 | 5 | 4 | 11 | 68 |
| 2002 | 10 | 7 | 6 | 6 | 6 | 5 | 4 | 0 | 0 | 0 | 1 | 0 | 45 |
| 2003 | 0 | 0 | 0 | 5 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 13 |
| 2004 | 6 | 0 | 0 | 1 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 15 |
| 2005 | 0 | 0 | 0 | 3 | 0 | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 10 |
| 2006 | 0 | 2 | 0 | 6 | 10 | 4 | 0 | 0 | 0 | 2 | 12 | 5 | 41 |
| 2007 | 5 | 0 | 3 | 2 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 17 |
| 2008 | 0 | 0 | 0 | 5 | 11 | 7 | 5 | 4 | 2 | 16 | 7 | 5 | 62 |
| 2009 | 7 | 7 | 12 | 15 | 16 | 8 | 6 | 6 | 5 | 7 | 4 | 8 | 101 |
| 2010 | 7 | 6 | 5 | 5 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 31 |
| 2011 | 6 | 6 | 8 | 10 | 6 | 5 | 9 | 7 | 4 | 8 | 9 | 5 | 83 |
| 2012 | 0 | 0 | 4 | 13 | 11 | 9 | 7 | 6 | 5 | 4 | 8 | 5 | 72 |
| 2013 | 5 | 7 | 17 | 18 | 11 | 7 | 4 | 4 | 5 | 4 | 7 | 9 | 98 |
| 2014 | 8 | 15 | 10 | 9 | 14 | 9 | 6 | 5 | 6 | 1 | 0 | 0 | 83 |
| 2015 | 1 | 0 | 4 | 5 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 17 |
| 2016 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 |
| 2017 | 11 | 9 | 9 | 15 | 13 | 12 | 3 | 4 | 5 | 1 | 0 | 1 | 83 |
| 2018 | 4 | 3 | 3 | 4 | 10 | 13 | 8 | 6 | 0 | 2 | 5 | 1 | 59 |
| 2019 | 0 | 0 | 2 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| 2020 | 0 | 0 | 1 | 8 | 5 | 4 | 0 | 0 | 0 | 0 | 1 | 3 | 22 |
| 2021 | 3 | 0 | 0 | 1 | 7 | 7 | 4 | 5 | 2 | 0 | 7 | 10 | 46 |
| 2022 | 3 | 2 | 0 | 1 | 9 | 8 | 9 | 6 | 3 | 4 | 16 | 3 | 64 |
| 2023 | 6 | 5 | 3 | 4 | 11 | 13 | 5 | 3 | 3 | 1 | 0 | 0 | 54 |
| 2024 | 0 | 0 | 0 | 4 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 6 | 19 |
| Monthly Total (%) | 7 | 6 | 9 | 14 | 15 | 12 | 7 | 5 | 5 | 5 | 7 | 8 | 100 |
We can also make a nice plot for printed reports.
Show code cell source
#make a pretty pdf of the table with great_tables
from great_tables import GT,html
dfGT = df.copy()
dfGT['Storm Year'] = df.index
# put the year column first
cols = dfGT.columns.tolist()
cols = cols[-1:] + cols[:-1]
dfGT = dfGT[cols]
dfGT.reset_index(drop=True, inplace=True)
# Create a GreatTable object
table = (GT(dfGT)
.fmt_number(columns=calendar.month_abbr[1:13], decimals=0)
.fmt_number(columns=['Annual'], decimals=0)
.tab_header(title = 'Days Exceeding 30 cm above MHHW', subtitle = station_name)
.data_color(domain = [0,20],
columns=calendar.month_abbr[1:13],
rows = list(range(len(dfGT)-1)),
palette=["white", "lightblue"])
.data_color(domain = [0,20],
columns=calendar.month_abbr[1:13],
rows = [-1],
palette=["white", "purple"])
.opt_table_outline(style='solid', width='3px', color='white')
)
# save table as png
tableName = station_name + '_flood_days_intra_annual.png'
savePath = os.path.join(output_dir, tableName)
# set size of table
# replace any commas or spaces with underscores
# savePath = savePath.replace(' ', '_')
table.save(savePath)
# Load Image
from IPython.display import Image
imgTable = Image(filename=savePath)
# Glue the image with a name
glue("imgTable", imgTable, display=False)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[27], line 2
1 #make a pretty pdf of the table with great_tables
----> 2 from great_tables import GT,html
4 dfGT = df.copy()
5 dfGT['Storm Year'] = df.index
ModuleNotFoundError: No module named 'great_tables'
# Define the decades for analysis
decades = [(1983, 1993), (1993, 2003), (2003, 2013), (2013, 2023)]
# Initialize lists to store results
total_flood_days_list = []
average_flood_days_per_year_list = []
percent_increase_days_per_year_list = [0] # First decade has no previous data for comparison
# Calculate statistics for each decade
for i, (start_year, end_year) in enumerate(decades):
# Filter the dataframe for the current decade
flood_days_decade = flood_days_per_year[(flood_days_per_year['year_storm'] >= start_year) & (flood_days_per_year['year_storm'] <= end_year)]
sum_flood_days_decade = flood_days_decade['flood_days_count'].sum()
avg_flood_days_decade = sum_flood_days_decade / len(flood_days_decade)
# Append results to lists
total_flood_days_list.append(sum_flood_days_decade)
average_flood_days_per_year_list.append(np.round(avg_flood_days_decade,0))
# Calculate percent increase for subsequent decades
if i > 0:
prev_avg_flood_days = average_flood_days_per_year_list[0]
percent_increase = np.round((avg_flood_days_decade - prev_avg_flood_days) / prev_avg_flood_days * 100, 1)
percent_increase_days_per_year_list.append(percent_increase)
# Create a dataframe for decadal statistics
decadal_stats = pd.DataFrame({
'decade': ['1983-1993', '1993-2003', '2003-2013', '2013-2023'],
'total_flood_days': total_flood_days_list,
'average_flood_days_per_year': average_flood_days_per_year_list,
'percent_increase_days_per_year': percent_increase_days_per_year_list
})
decadal_stats
| decade | total_flood_days | average_flood_days_per_year | percent_increase_days_per_year | |
|---|---|---|---|---|
| 0 | 1983-1993 | 194 | 18.0 | 0.0 |
| 1 | 1993-2003 | 350 | 32.0 | 76.8 |
| 2 | 2003-2013 | 524 | 48.0 | 164.6 |
| 3 | 2013-2023 | 528 | 48.0 | 166.7 |
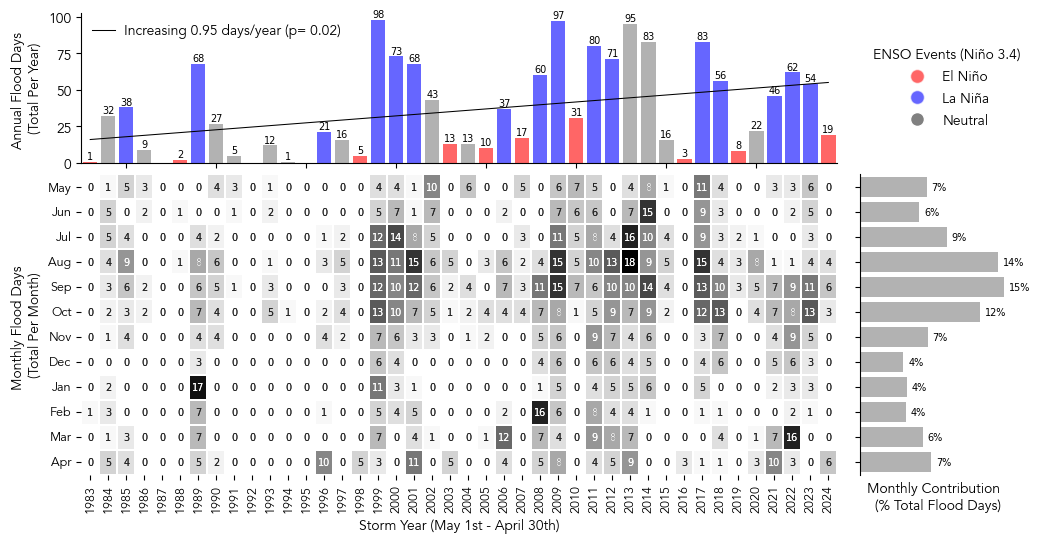
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
# Create a figure and shared axis layout
fig, axs = plt.subplots(2,2,figsize=(12,6), gridspec_kw={"height_ratios": [1, 2], "hspace": 0.05, "width_ratios":[5,1], "wspace": 0.05})
# put stuff in the figure
# Create heatmap on the left
sns.heatmap(df.iloc[:-1, :-1].T, cmap="Blues", annot=True, fmt=".0f", linewidths=0.2,
ax=axs[1,0],cbar=False, annot_kws={"size": 7})
# heatmap
sns.heatmap(df.iloc[:-1, :-1].T, cmap="Grays", annot=True, fmt=".0f", linewidths=0.2,
ax=axs[1,0],cbar=False, annot_kws={"size": 7})
axs[1,0].set_ylabel("Monthly Flood Days \n(Total Per Month)")
axs[1,0].set_xlabel("Storm Year (May 1st - April 30th)")
axs[1,0].tick_params(axis='y', labelsize=9)
axs[1,0].tick_params(axis='x', labelsize=9)
# map oni_year to colors using enso (we will keep this for now for possible future use)
# oni_colors = ['red' if val > 0.5 else 'blue' if val < -0.5 else 'gray' for val in flood_days_per_year['ONI']]
# color by enso_event
enso_colors = ['red' if val == 'El Niño' else 'blue' if val == 'La Niña' else 'gray' for val in flood_days_per_year['event']]
# yearly totals
ax2 = axs[0,0]
# set ax2 x-axis to be the same as ax1
bars = ax2.bar(df.index[0:-1], df["Annual"][0:-1], color=enso_colors, alpha=0.6, label="Annual Flood Days")
ax2.set_xlim(df.index[0]-0.5, df.index[-2]+0.5)
ax2.set_ylabel("Annual Flood Days \n(Total Per Year)")
# Add annotations to bars
for bar in bars:
height = bar.get_height()
if height > 0:
ax2.text(bar.get_x() + bar.get_width() / 2, height + 0.5, f"{int(height)}",
ha='center', va='bottom', fontsize=7, color="black")
# remove top and right boudaries
ax2.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
# bar plot of monthly totals
ax3 = axs[1,1]
barMonth = ax3.barh(range(1, 13), df.iloc[-1, :-1][::-1], color="gray", alpha=0.6, label="Mean Yearly Flood Days")
ax3.set_yticks(range(1, 13))
ax3.set_ylim(0.5, 12.5)
# remove y-axis labels
ax3.set_yticklabels([])
#remove x-axis ticks and labels
ax3.set_xticks([])
ax3.set_xlabel("Monthly Contribution \n (% Total Flood Days)")
#remove top and right boudaries
for spine in ax3.spines.values():
spine.set_visible(False)
ax3.spines['left'].set_visible(True)
# add annotations to bars
for bar in barMonth:
width = bar.get_width()
if width > 0:
ax3.text(width + 0.5, bar.get_y() + bar.get_height() / 2, f"{int(width)}%",
ha='left', va='center', fontsize=7, color="black")
# ax[0,1] defines the oni_color legend
ax4 = axs[0,1]
ax4.axis('off')
# create a legend for the oni_colors
legend_elements = [plt.Line2D([0], [0], marker='o', color='w', label='El Niño', markerfacecolor='red', markersize=10, alpha=0.6),
plt.Line2D([0], [0], marker='o', color='w', label='La Niña', markerfacecolor='blue', markersize=10, alpha=0.6),
plt.Line2D([0], [0], marker='o', color='w', label='Neutral', markerfacecolor='gray', markersize=10)]
legend = ax4.legend(handles=legend_elements, loc='center left', title='ENSO Events (Niño 3.4)', frameon=False)
legend_title = legend.get_title()
legend_title.set_fontsize('10')
legend_title.set_fontname('Avenir')
legend_texts = legend.get_texts()
for text in legend_texts:
text.set_fontname('Avenir')
text.set_fontsize('10')
# add trendline to the yearly totals
x = df.index[0:-1].astype(int).values
y = df["Annual"][0:-1].values
slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
trendDays = intercept + slope * x
trendLabel = 'Increasing {:.2f} days/year (p= {:.2f})'.format(slope,p_value)
if p_value < 0.05:
linestyleTrend = '-'
else:
linestyleTrend = '--'
ax2.plot(x, trendDays, color='black', linestyle=linestyleTrend, label=trendLabel, linewidth=0.75)
# add another legend for the trendline
# Add a new legend only for the trendline
legendTrend = ax2.legend([plt.Line2D([0], [0], color='black', linestyle=linestyleTrend, linewidth=0.75)],
[trendLabel], loc='upper left', fontsize='small', frameon=False)
legendTrend_texts = legendTrend.get_texts()
for text in legendTrend_texts:
text.set_fontname('Avenir')
text.set_fontsize('10')
# set all fonts in the figure to Avenir
# Iterate over all axes in the figure
for ax in axs.flat:
# Change font for title, axis labels, and tick labels
for item in ([ax.title, ax.xaxis.label, ax.yaxis.label] +
ax.get_xticklabels() + ax.get_yticklabels()):
item.set_fontname('Avenir')
#save the figure
fig.savefig(output_dir / 'SL_FloodFrequency_threshold_counts_heatmap.png', bbox_inches='tight')
plt.savefig(op.join(path_figs, 'F11_Minor_flood_matrix.png'), dpi=300, bbox_inches='tight')
Create a map#
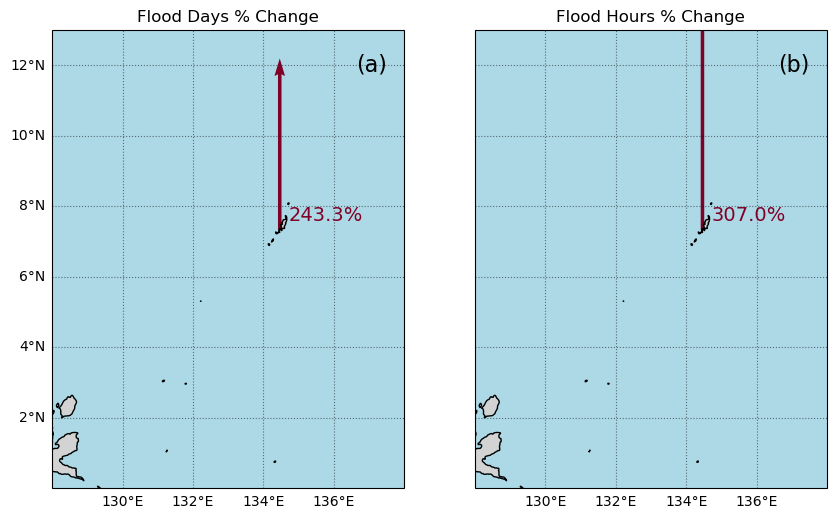
In this next section we’ll create a map with an icon depicting the percent change over the Period of Record at the tide station/s, for both Frequency and Duration.
The following cell defines a function to add arrow icons denoting percent change.
Show code cell source
# make a function for adding the arrows
def add_arrow(ax, lat,lon,percent_change,crs, vmin, vmax):
# make colormap of percent change
adjusted_heatmap_palette = sns.color_palette("YlOrRd", as_cmap=True)
# Prepare data for quiver plot
U = np.zeros_like(lon) # Dummy U component (no horizontal movement)
V = percent_change/100 # V component scaled by percent change
arrow_scale = 5 # Adjust as necessary for arrow size
arrow_width = 0.01 # Adjust for desired arrow thickness
# Quiver plot
q = ax.quiver(lon,lat, U, V, transform=crs, scale=arrow_scale,
color=adjusted_heatmap_palette(percent_change / vmax),
cmap=adjusted_heatmap_palette, clim=(vmin, vmax), width=arrow_width)
And here is our final plotting code:
Show code cell source
xlims = [128, 138]
ylims = [0, 13]
vmin, vmax = 0,100
# fig, ax, crs,cmap = plot_map(vmin,vmax,xlims,ylims)
crs = ccrs.PlateCarree()
fig, axs = plt.subplots(1, 2, figsize=(10, 6), subplot_kw={'projection': crs})
for i, ax in enumerate(axs):
ax.set_xlim(xlims)
ax.set_ylim(ylims)
ax.coastlines()
# Fill in water
ax.add_feature(cfeature.LAND, color='lightgrey')
# add a) b) labels
ax.text(0.95, 0.95, f'({chr(97 + i)})',
horizontalalignment='right', verticalalignment='top', transform=ax.transAxes,
fontsize=16)
ax.add_feature(cfeature.OCEAN, color='lightblue')
# add map grid
gl = ax.gridlines(draw_labels=True, linestyle=':', color='black',
alpha=0.5,xlocs=ax.get_xticks(),ylocs=ax.get_yticks())
#remove labels from top and right axes
gl.top_labels = False
gl.right_labels = False
if ax == axs[1]:
gl.left_labels = False
add_arrow(axs[0], rsl['lat'], rsl['lon'], percent_change_days, crs, vmin, vmax)
add_arrow(axs[1], rsl['lat'], rsl['lon'], percent_change_hours, crs, vmin, vmax)
# Put titles on the subplots
axs[0].set_title('Flood Days % Change')
axs[1].set_title('Flood Hours % Change')
# Add text for percent change
for i in range(len(rsl['lon'])):
axs[0].text(rsl['lon'][i] + 0.25, rsl['lat'][i] + 0.25, '{:.1f}%'.format(percent_change_days), fontsize=14,
color=adjusted_heatmap_palette(percent_change_days / vmax))
axs[1].text(rsl['lon'][i] + 0.25, rsl['lat'][i] + 0.25, '{:.1f}%'.format(percent_change_hours), fontsize=14,
color=adjusted_heatmap_palette(percent_change_hours / vmax))
# glue("mag_fig", fig, display=False)
# Save the figure
output_file_path = output_dir / 'SL_FloodFrequency_map.png'
fig.savefig(output_file_path, dpi=300, bbox_inches='tight')