Estimated Phytoplankton Size#
Show code cell source
import warnings
warnings.filterwarnings("ignore")
import os
import os.path as op
import sys
import pandas as pd
import numpy as np
import xarray as xr
import geopandas as gpd
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
sys.path.append("../../../../indicators_setup")
from ind_setup.plotting_int import plot_timeseries_interactive
from ind_setup.plotting import plot_base_map, plot_map_subplots, plot_bar_probs
from ind_setup.core import fontsize
sys.path.append("../../../functions")
from data_downloaders import download_ERDDAP_data
Define area of interest
#Area of interest
lon_range = [129.4088, 137.0541]
lat_range = [1.5214, 11.6587]
EEZ shapefile
shp_f = op.join(os.getcwd(), '..', '..','..', 'data/Palau_EEZ/pw_eez_pol_april2022.shp')
shp_eez = gpd.read_file(shp_f)
Download Data#
DATASET: https://oceanwatch.pifsc.noaa.gov/erddap/info/md50_exp/index.html
update_data = False
path_data = "../../../data"
path_figs = "../../../matrix_cc/figures"
Show code cell source
base_url = 'https://oceanwatch.pifsc.noaa.gov/erddap/griddap/md50_exp.csv'
dataset_id = 'MD50'
if update_data:
date_ini = '1998-01-01T00:00:00Z'
date_end = '2023-12-01T00:00:00Z'
data = download_ERDDAP_data(base_url, dataset_id, date_ini, date_end, lon_range, lat_range)
data_xr = data.set_index(['latitude', 'longitude', 'time']).to_xarray()
data_xr['time'] = pd.to_datetime(data_xr.time)
data_xr = data_xr.coarsen(longitude=2, latitude=2, boundary = 'pad').mean()
data_xr.to_netcdf(op.join(path_data, f'griddap_{dataset_id}.nc'))
else:
data_xr = xr.open_dataset(op.join(path_data, f'griddap_{dataset_id}.nc'))
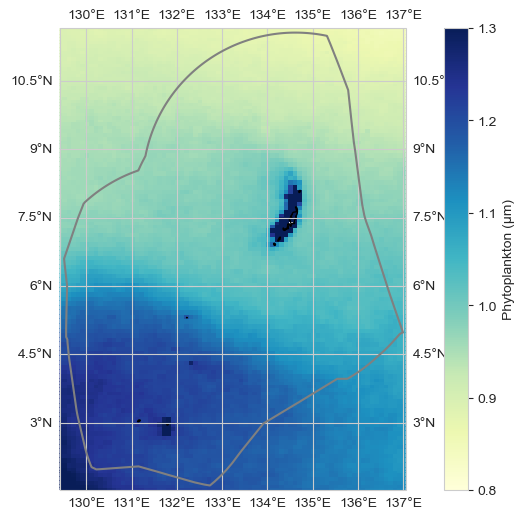
ax = plot_base_map(shp_eez = shp_eez, figsize = [10, 6])
im = ax.pcolor(data_xr.longitude, data_xr.latitude, data_xr.mean(dim='time')[dataset_id], transform=ccrs.PlateCarree(),
cmap = 'YlGnBu', vmin = 0.8, vmax = 1.3)
ax.set_extent([lon_range[0], lon_range[1], lat_range[0], lat_range[1]], crs=ccrs.PlateCarree())
plt.colorbar(im, ax=ax, label='Phytoplankton (µm)')
plt.savefig(op.join(path_figs, 'F16_phytoplankton_mean_map.png'), dpi=300, bbox_inches='tight')
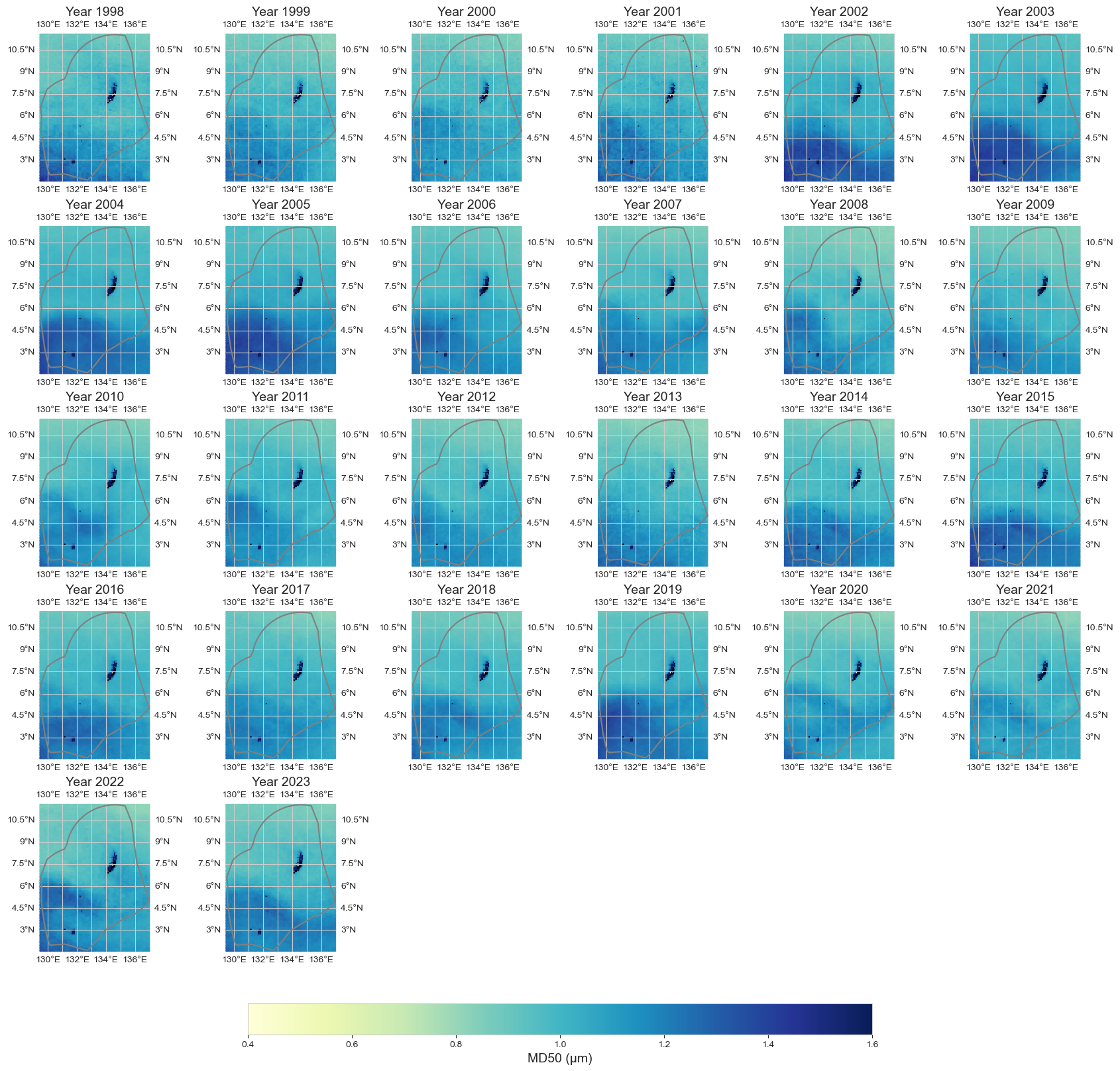
data_y = data_xr.resample(time='1YE').mean()
plot_map_subplots(data_y, 'MD50', shp_eez = shp_eez, cmap = 'YlGnBu', vmin = 0.4, vmax = 1.6, cbar = 1)

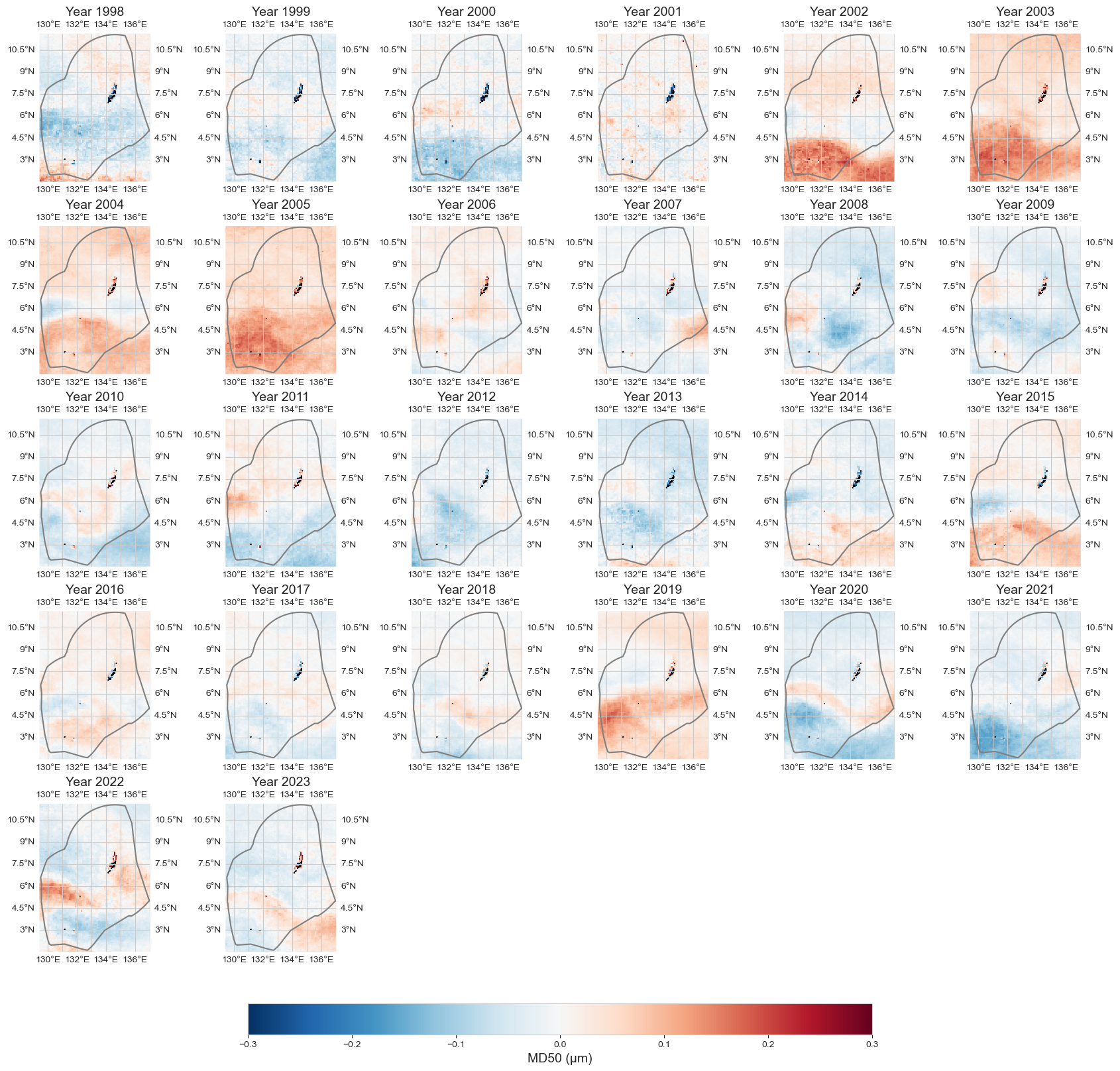
data_an = data_y - data_xr.mean(dim='time')
plot_map_subplots(data_an, dataset_id, shp_eez = shp_eez, cmap='RdBu_r', vmin=-.3, vmax=.3, cbar = 1)

Mean Area#
dict_plot = [{'data' : data_xr.mean(dim = ['longitude', 'latitude']).to_dataframe(),
'var' : dataset_id, 'ax' : 1, 'label' : 'Median Phytoplankton Size - MEAN AREA'},]
fig = plot_timeseries_interactive(dict_plot, trendline=True, scatter_dict = None, figsize = (25, 12));
fig.write_html(op.join(path_figs, 'F16_phytoplankton_mean_trend.html'), include_plotlyjs="cdn")
Given point#
loc = [7.35, 134.48]
dict_plot = [{'data' : data_xr.sel(longitude=loc[1], latitude=loc[0], method='nearest').to_dataframe(),
'var' : dataset_id, 'ax' : 1, 'label' : f'Median Phytoplankton Size at [{loc[0]}, {loc[1]}]'},]
ax = plot_base_map(shp_eez = shp_eez, figsize = [10, 6])
ax.set_extent([lon_range[0], lon_range[1], lat_range[0], lat_range[1]], crs=ccrs.PlateCarree())
ax.plot(loc[1], loc[0], '*', markersize = 12, color = 'royalblue', transform=ccrs.PlateCarree(), label = 'Location Analysis')
ax.legend()
<matplotlib.legend.Legend at 0x185e7c590>
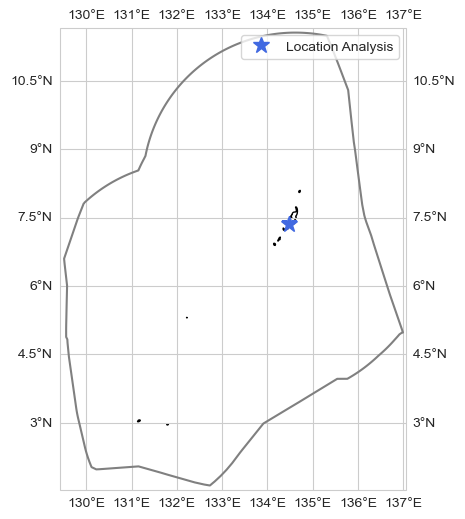
fig = plot_timeseries_interactive(dict_plot, trendline=True, scatter_dict = None, figsize = (25, 12));
